The Bond object
The Bond object is used to draw bond for Batoms. It has a settings attribute (BondSettings object), which stores and sets all parameters related to bonds. Possible keywords for settings are: symbol1, symbol2, min, max, search, polyhedra, color1, color2, bondlinewidth, style, material_style.
>>> from ase.build import molecule
>>> from batoms import Batoms
>>> ch4 = Batoms('ch4', from_ase = molecule('CH4'))
>>> ch4.model_style = 2

You can print the default bond.settings by:
>>> ch4.bond.settings
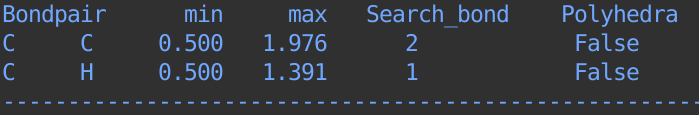
By defaut, we use default radius (ase.data.covalent_radii) for every atoms, and the maximum bondlength is the sum of two radius and then scaled by a default cutoff (1.3). The minimum bondlength is 0.5.
Style
One set the bond style for a bond pair by:
>>> ch4.bond.settings[('C', 'H')].style = '3'
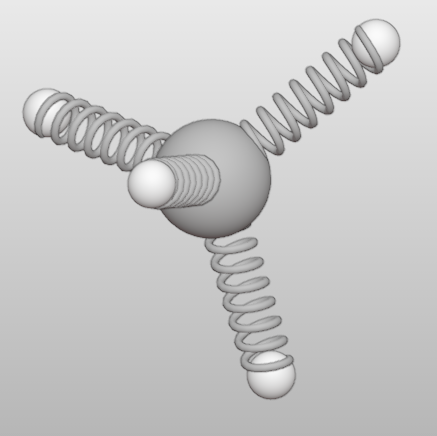
One set the bond style for a bond by (Here is the first bond):
>>> ch4.bonds[0].style = '3'

Here, four bond models are supported.
|
|
|
|
|
|
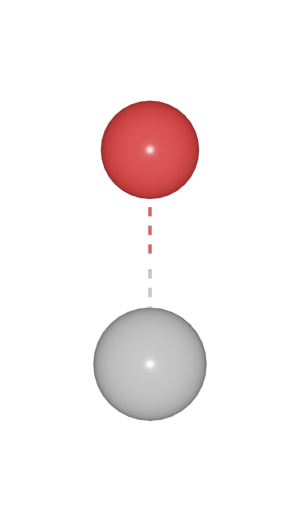
|
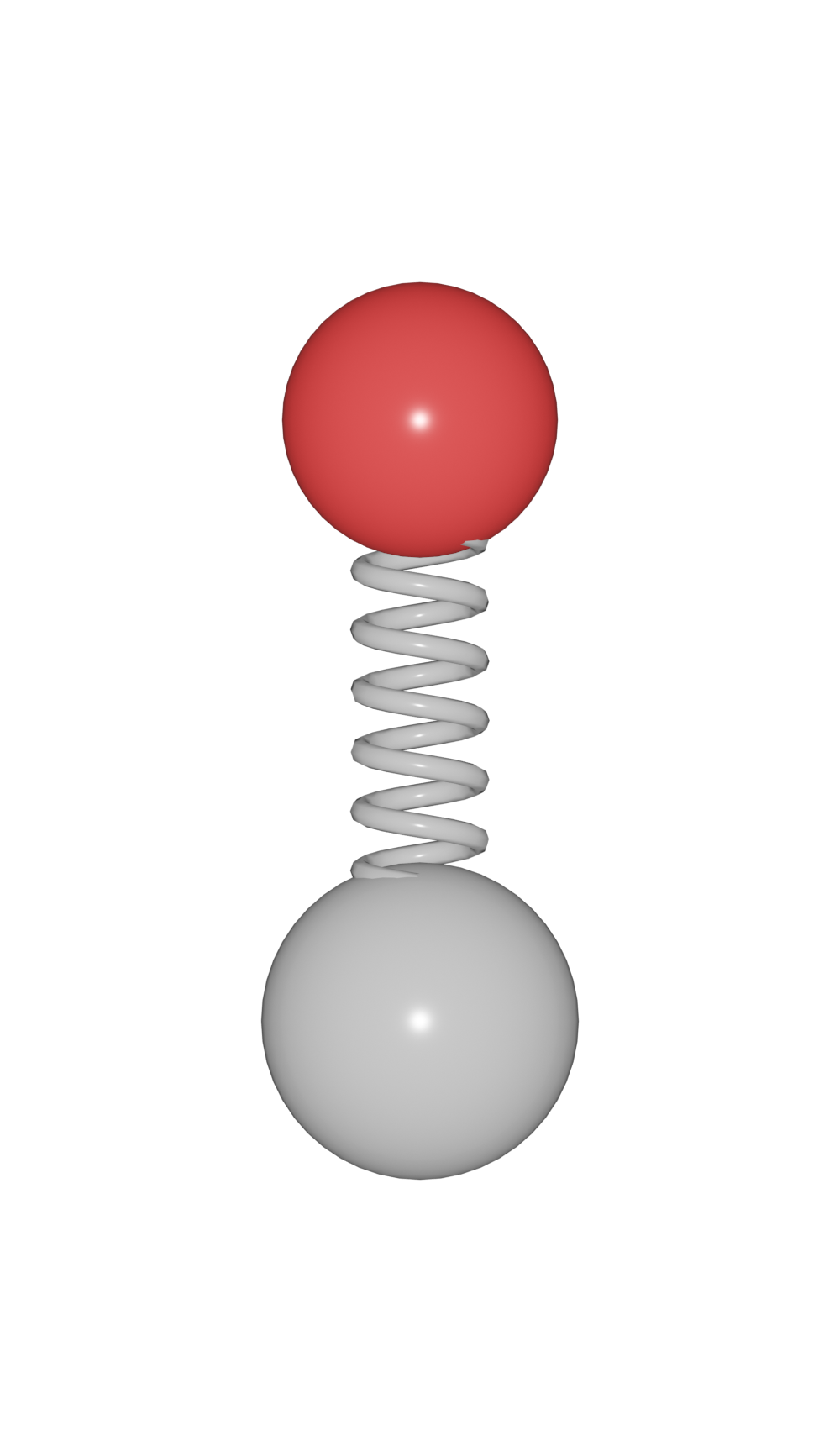
|
Polyhedra
One can change setting for a bond pair. For example, to build up coordination polyhedra, the value for polyhedra should be set to True:
>>> ch4.bond.settings[('C', 'H')].polyhedra = True
>>> ch4.model_style = 2
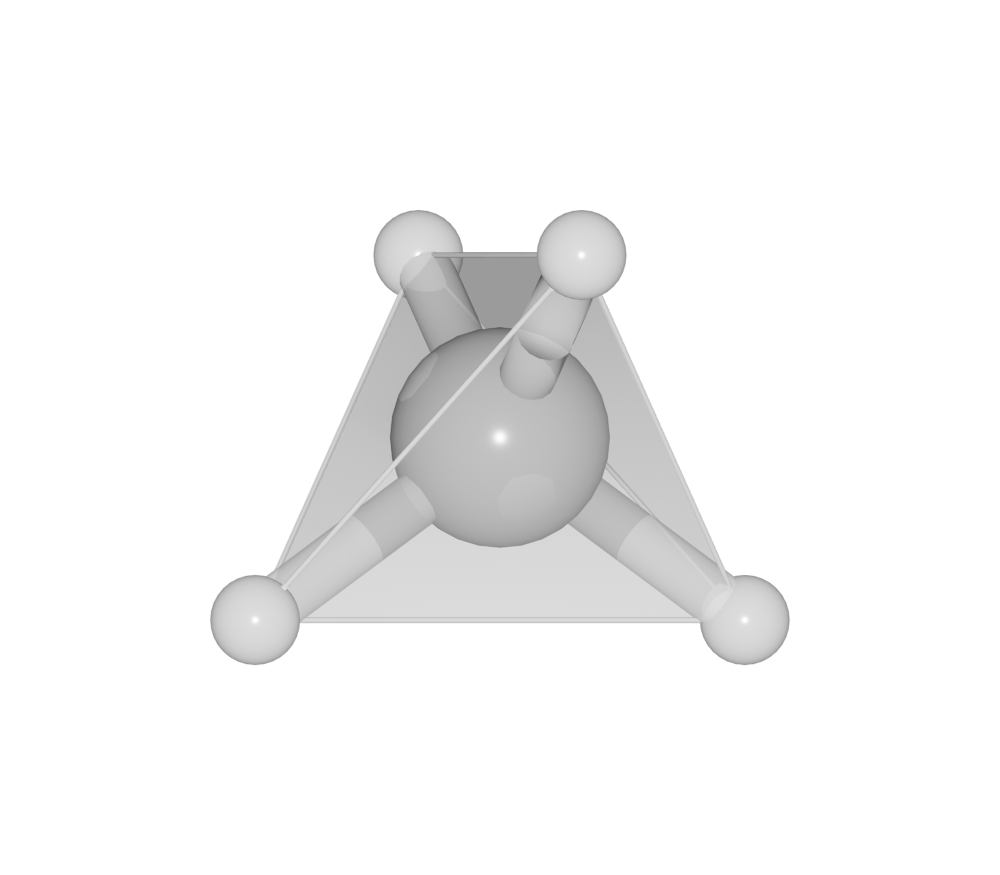
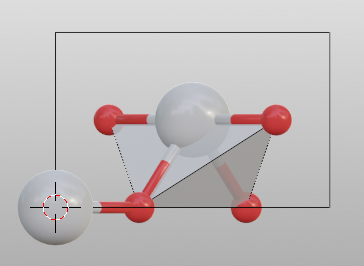
Search bond mode
0Do not search atoms beyond the boundary
1Search additional atoms if species1 is included in the boundary
2Search bonded atoms of species1 or species2 recursively. This mode is the used for searching molecules.
To change setting for search by:
>>> tio2.bond.settings[('Ti', 'O')].search = 0
>>> tio2.update_boundary()
>>> tio2.model_style = 2
Color
One can print the default color by:
>>> ch4.bond.settings[('C', 'H')].color1[:]
One can change color for a bond pair.
>>> ch4.bond.settings[('C', 'H')].color1 = [0.8, 0.1, 0.3, 0.5]
>>> ch4.bond.settings[('C', 'H')].color2 = [0.1, 0.3, 0.2, 1.0]
>>> ch4.model_style = 1
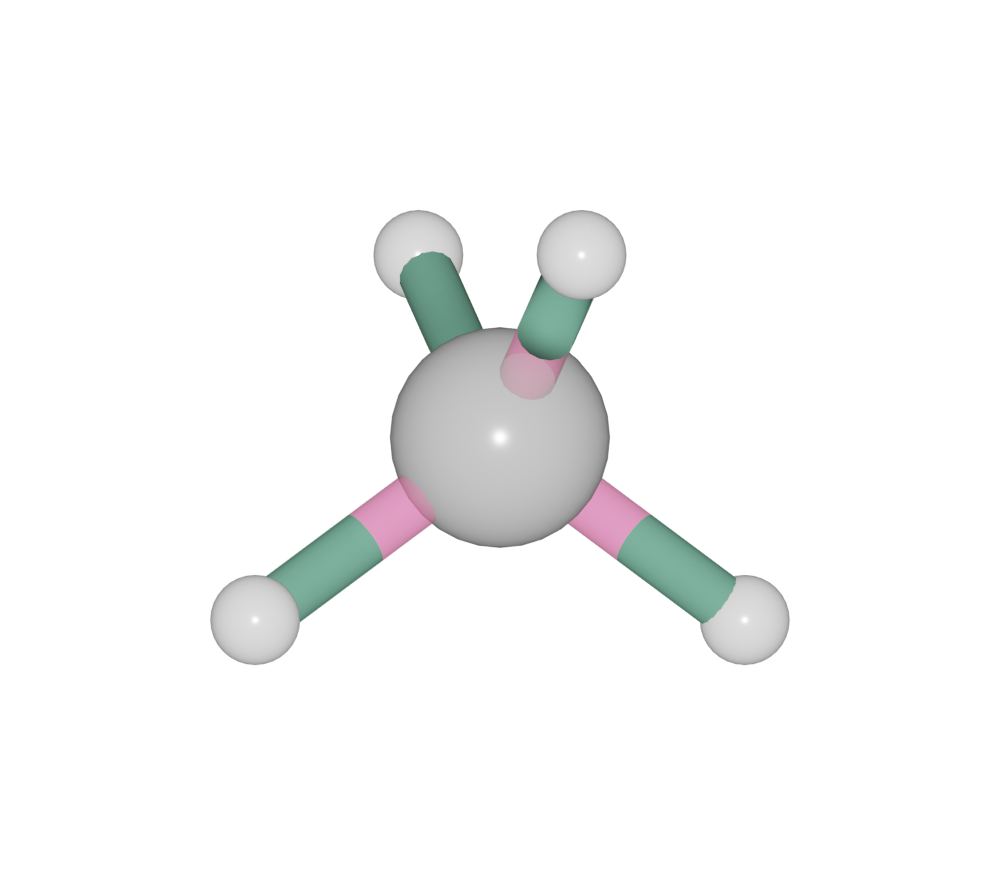
High order bond
One can set bond order for each bond:
>>> from ase.build import molecule
>>> from batoms import Batoms
>>> c6h6 = Batoms("c6h6", from_ase = molecule("C6H6"))
>>> c6h6.model_style = 1
>>> c6h6.bonds[0].order = 2
>>> c6h6.bonds[5].order = 2
>>> c6h6.bonds[9].order = 2
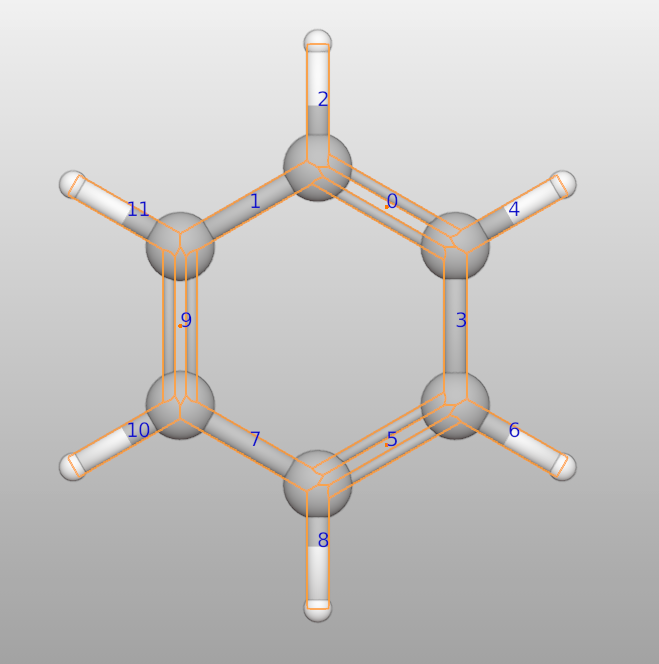
Or one can set the bond order automaticaly based on pybel:
>>> c6h6.bonds.bond_order_auto_set()