Bond and Polyhedra
The Bondsettings object controls various settings such as the bondlength and the polyhedra.
One can set model_style to draw the bond and polyhedra.
>>> from batoms.bio import read
>>> tio2 = read("tio2.cif")
>>> tio2.model_style = 1
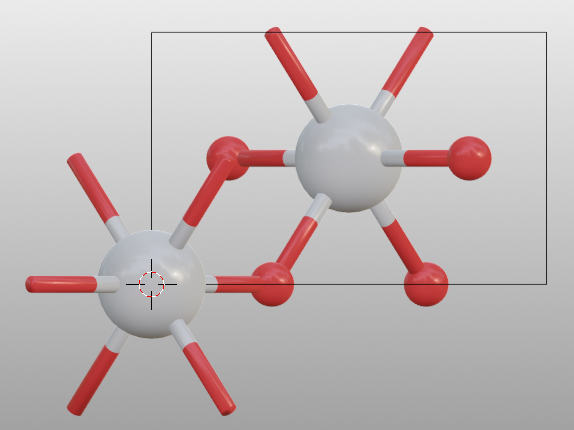
You can print the default bondsetting by:
>>> tio2.bond.settings
Bondpair min max Search_bond Polyhedra style
Ti-O 0.000 2.938 1 True 1
O-O 0.000 1.716 1 False 1
------------------------------------------------------------
To build up coordination polyhedra, set model_style to 2.
>>> tio2.model_style = 2
>>> # show atoms
>>> tio2.bond.show_search = True
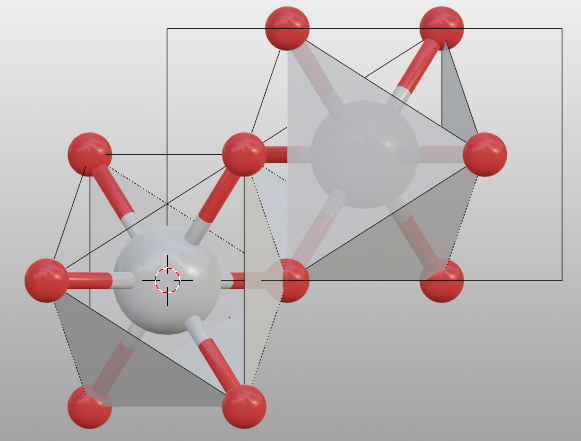
One can remove or add a bond pair by:
>>> tio2.bond.settings.remove(("Ti", "O"))
>>> tio2.bond.settings.add(("Ti", "O"))
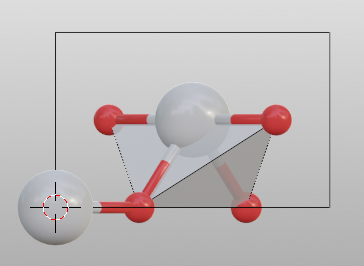
Search bond mode
Do not search atoms beyond the boundary. The value for
searchshould be set to 0.
>>> tio2.bond.settings[("Ti", "O")].search = 0
>>> tio2.boundary.update()
>>> tio2.model_style = 2
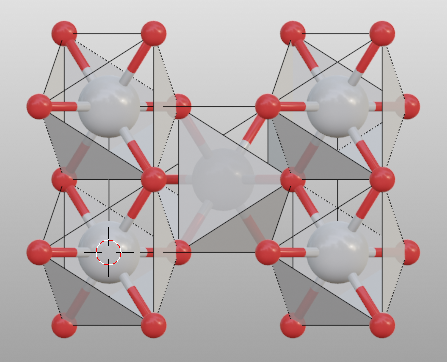
Search additional atoms if species1 is included in the boundary, the value for
searchshould be set to >0. To change setting for a bond pair by.
>>> tio2.boundary = 0.01
>>> tio2.bond.settings[("Ti", "O")].search = 1
>>> tio2.model_style = 2
>>> # show atoms
>>> tio2.bond.show_search = True
Search bonded atoms of species1 or species2 recursively. This mode is the used for searching molecules.
>>> from batoms.bio import read
>>> mol = read("urea.cif")
>>> mol.boundary = 0.01
>>> mol.model_style = 1
>>> mol.bond.show_search = True
>>> mol.get_image([1, -0.3, 0.1], engine = "eevee", output = "bondsetting_search_molecule.png")
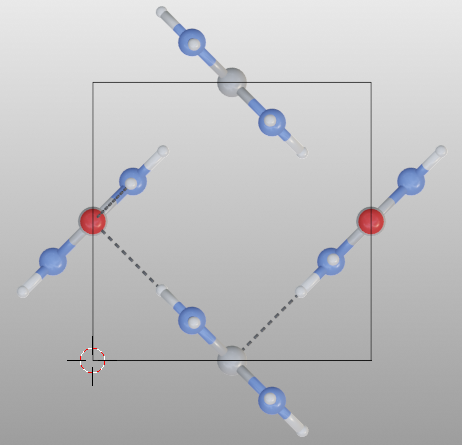
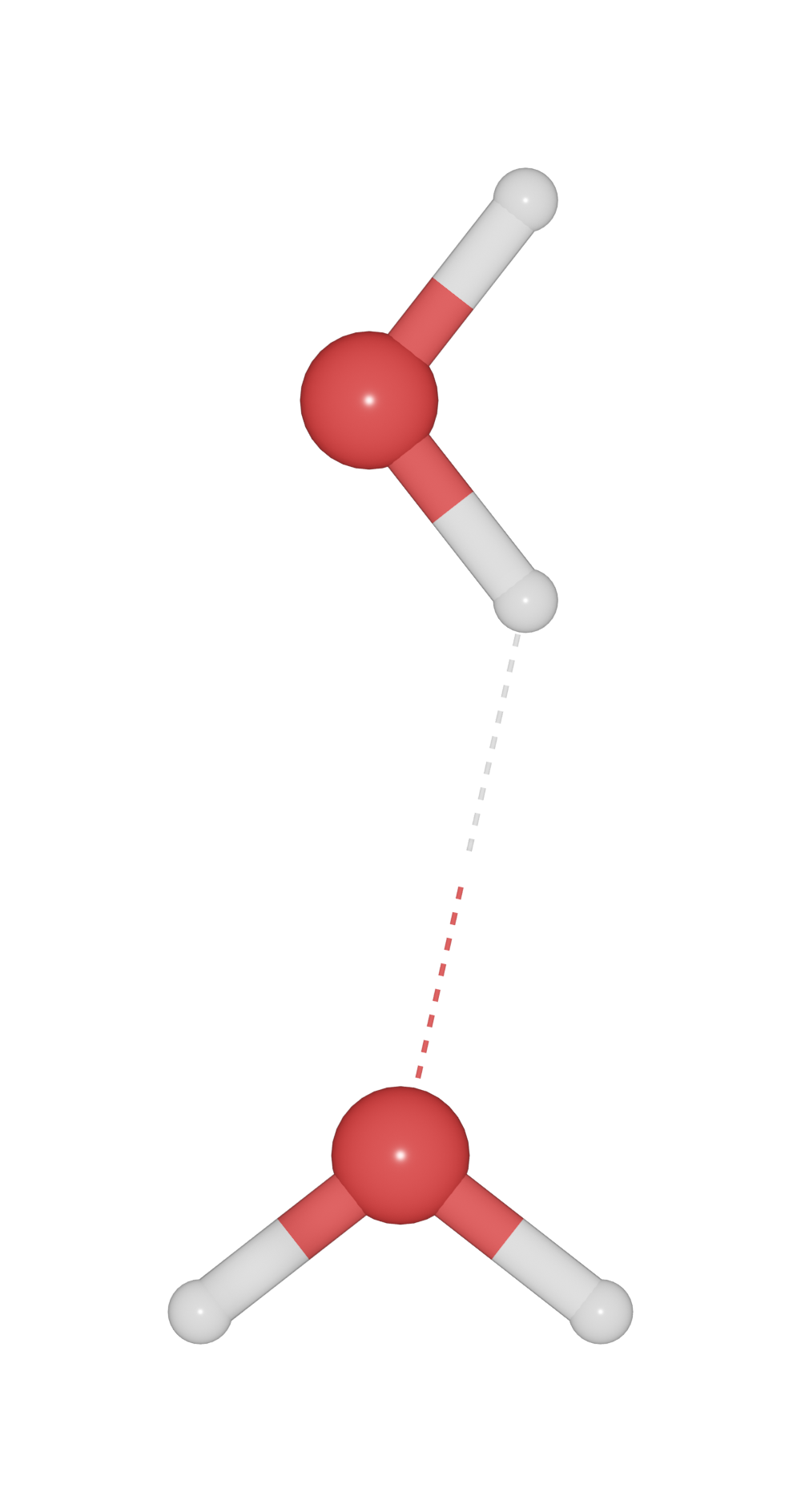
Hydrogen bond
To build up hydrogen bond for X-H -- Y. Set the minimum and maximum distances of H-Y, and set the bondlinewdith to a small value. Such as H-O and H-N bond.
>>> h2o.bond.settings[("H", "O")].min = 2.0
>>> h2o.bond.settings[("H", "O")].max = 3.0
High order bond
One can set bond order for each bond:
>>> from ase.build import molecule
>>> from batoms import Batoms
>>> c6h6 = Batoms("c6h6", from_ase = molecule("C6H6"))
>>> c6h6.model_style = 1
>>> c6h6.bonds[0].order = 2
>>> c6h6.bonds[5].order = 2
>>> c6h6.bonds[9].order = 2
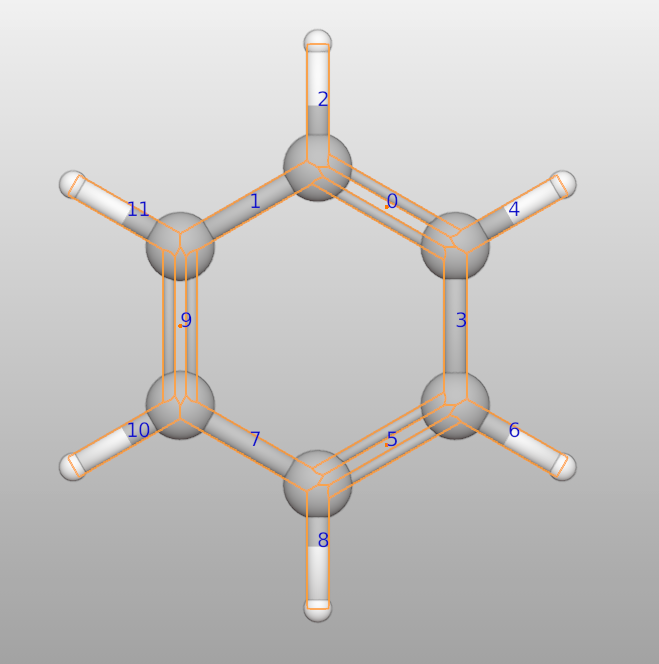
Or one can set the bond order automaticaly based on pybel:
>>> c6h6.bond.bond_order_auto_set()