Boundary mode
The boundary key in Batoms is used to search atoms outside the cell. The default value is [0, 0, 0], thus no atoms outside the cell will be search.
>>> from ase.build import bulk
>>> from batoms.batoms import Batoms
>>> au = bulk("Au", "fcc", cubic=True)
>>> au = Batoms(label = "au", from_ase = au)
>>> au.get_image(viewport = [1, -0.3, 0.1], output = "au.png")

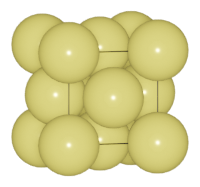
Then we change boundary to [0.02, 0.02, 0.02]:
>>> au.boundary = [0.02, 0.02, 0.02]
In this case, atoms in the range of [-0.02, 1.02] of the unit cell will be searched. Because the structure is periodic, so we get more Au atoms in the boundary (corner and face).
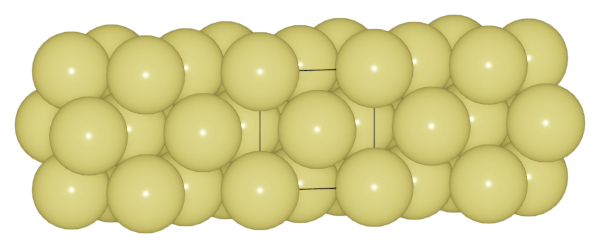
>>> au.boundary = [0.02, 2, 0.02]
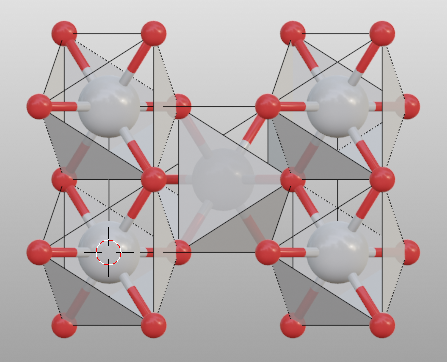
Search additional atoms if species1 is included in the boundary:
>>> from batoms.batoms import Batoms
>>> from batoms.bio import read
>>> tio2 = read("tio2.cif")
>>> tio2.boundary = 0.01
>>> tio2.model_style = 2